Astrochemical Forecasting
with Machine Learning
- Kelvin Lee,
- Jacqueline Patterson,
- Andrew Burkhardt,
- Michael McCarthy,
- Brett McGuire
ISMS 2021—Talk RC03
Molecules in space
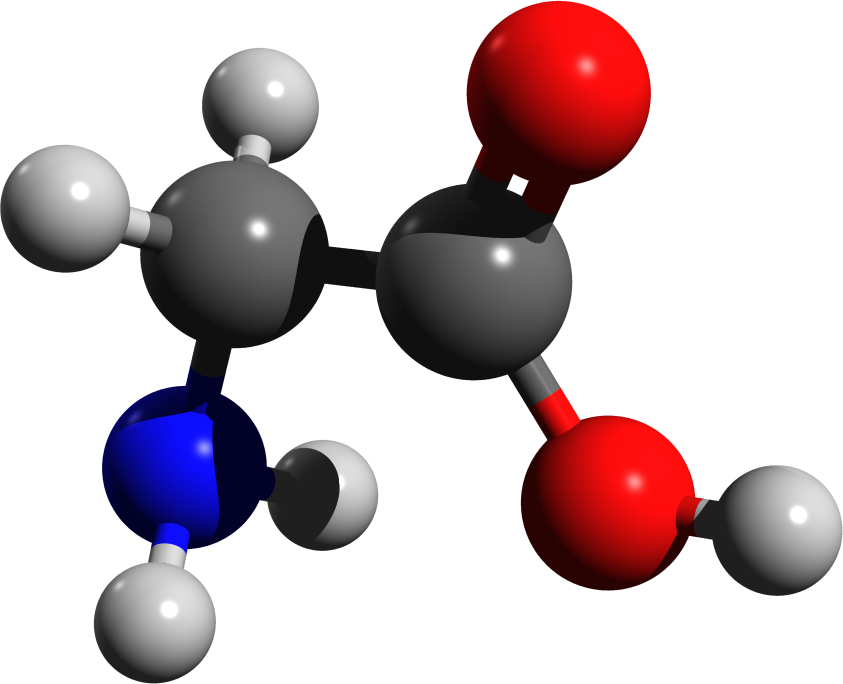
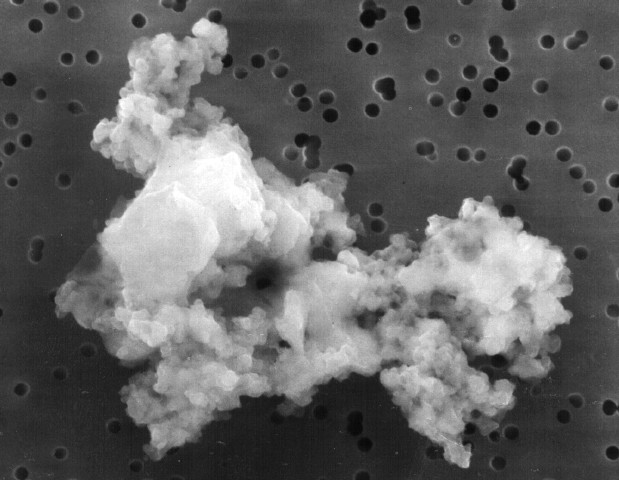
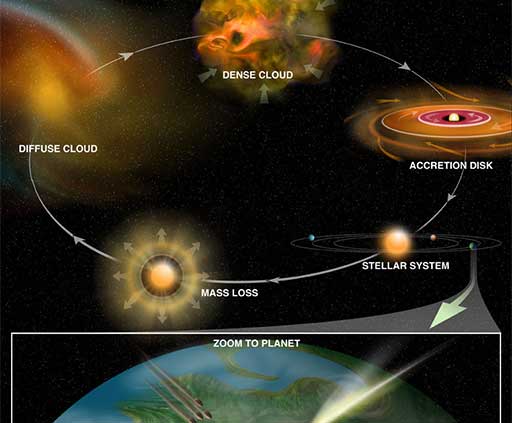
From origins of life to star formation
Pushing the boundaries of molecular complexity
See talk RC06 by Brett McGuire for the most up to date numbers!
What's next for astrochemistry?

Cheminformatics
High-throughput detection of desirable molecules from millions of candidates
Combine with machine learning to recommend molecules for study
Machine representations of molecules

Hand-picked features
Unsupervised representation learning
mol2vec
- Unsupervised language model adapted for molecule strings.
- SMILES encoding transformed into k-neighborhood chemical environment
- Generate a dictionary mapping between environments and vectors
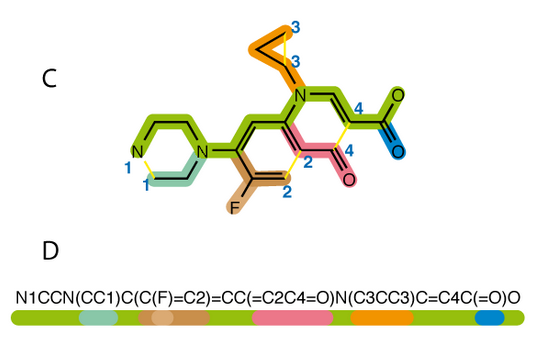
Molecule vector representations
Distance and similarity measures in Euclidean space
$\cos(\theta) = \frac{\mathbf{A} \cdot \mathbf{B}}{\vert \mathbf{A} \vert \vert \mathbf{B} \vert}$
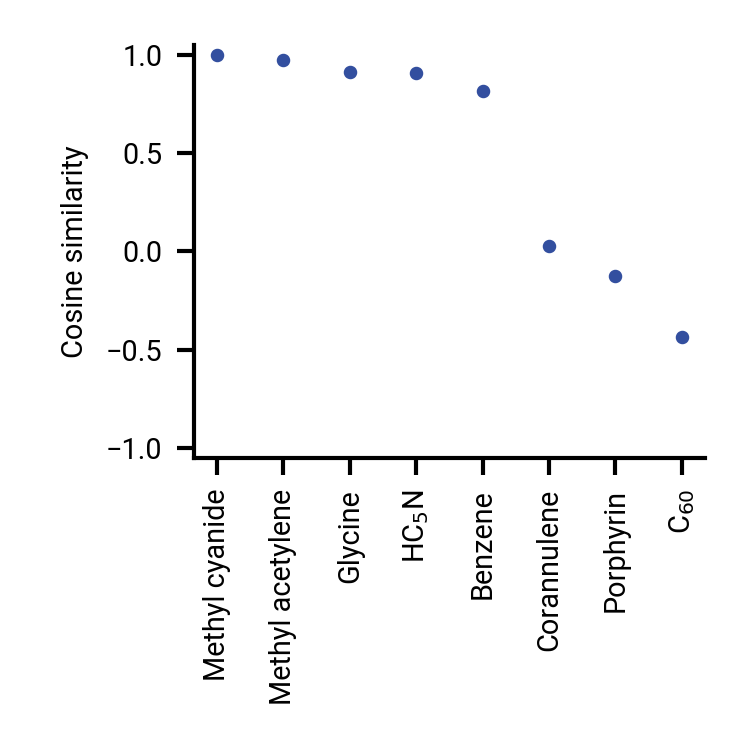
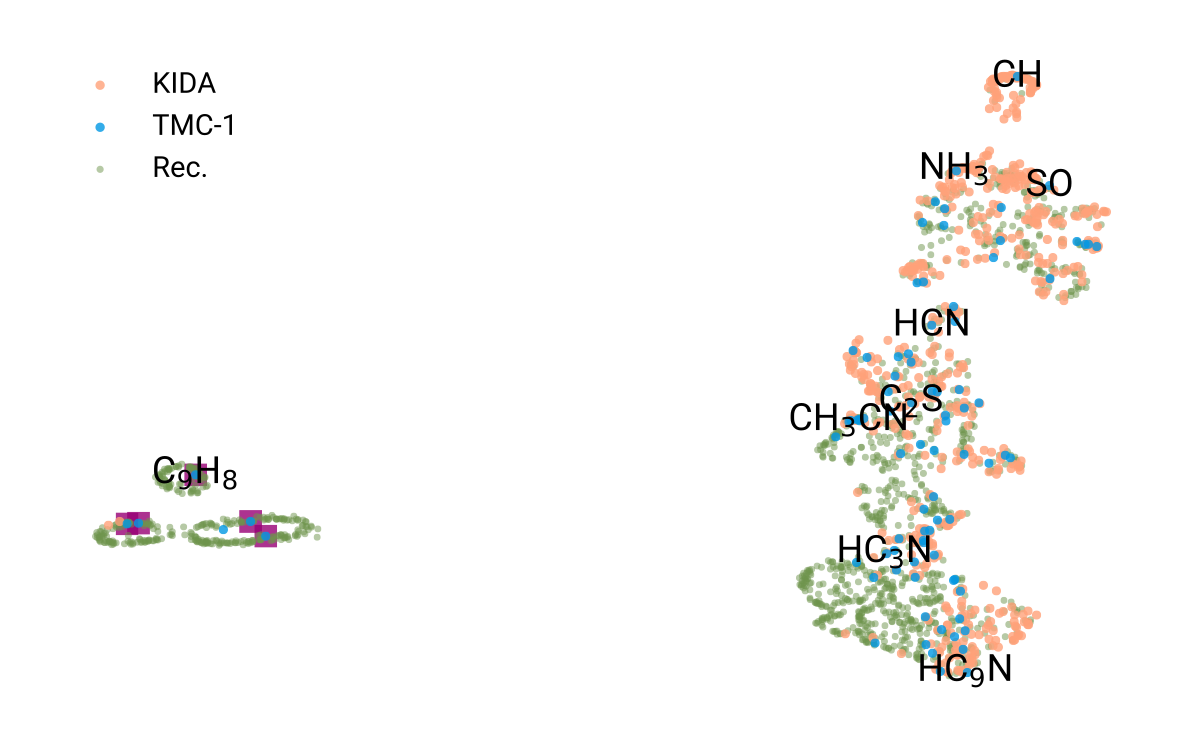
Molecule vector representations
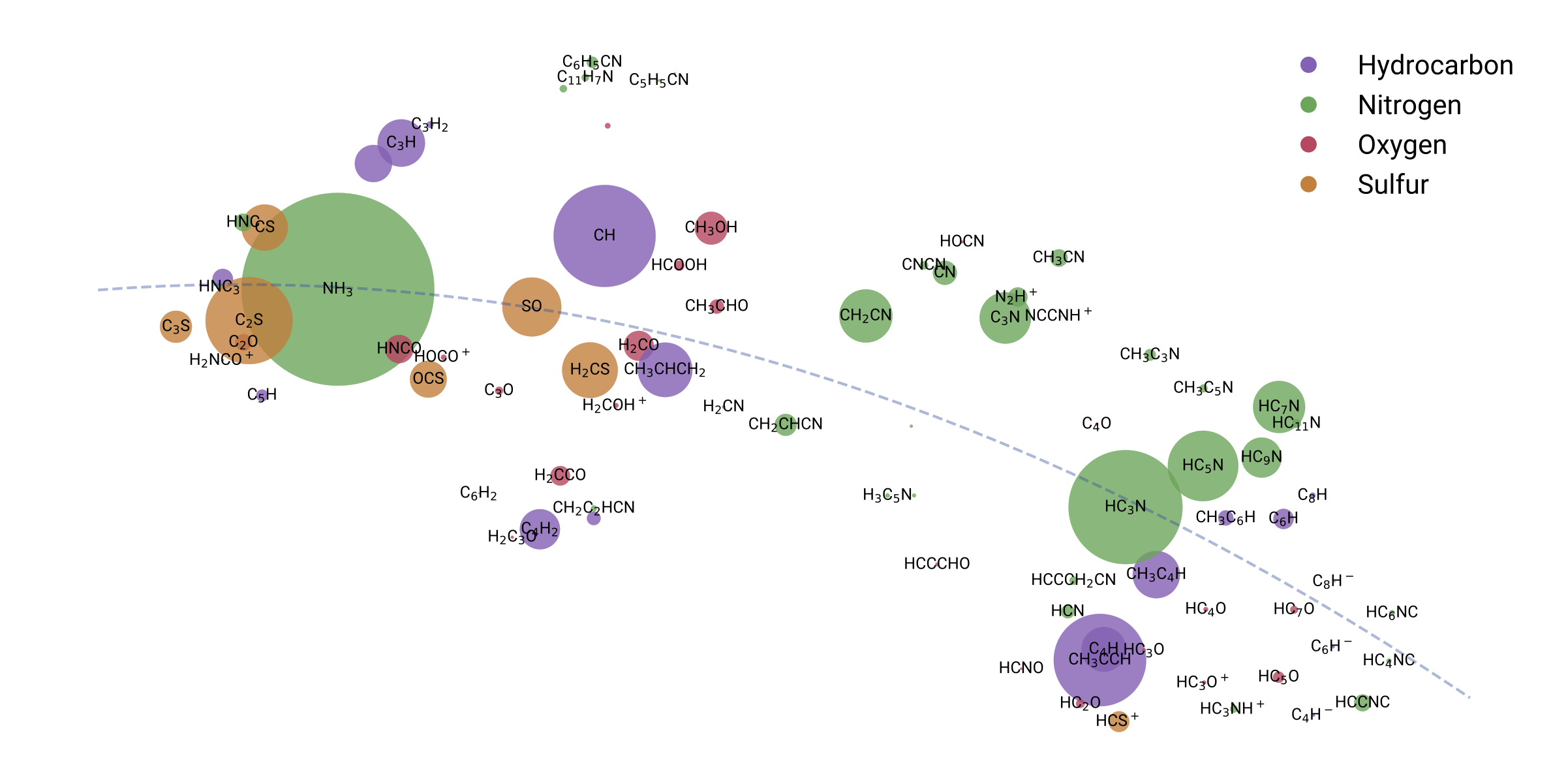
Visualization of chemical inventory clustering and trends
| Source | Number of entries |
| ZINC | 3,862,980 |
| PubChem A | 2,444,441 |
| PCBA | 437,929 |
| QM9 | 133,885 |
| NASA PAH database | 3,139 |
| KIDA | 578 |
| TMC-1 inventory | 88 |
Total of 3,316,454 unique molecules used for mol2vec training
455,461 chosen from unsupervised clustering based on proximity to TMC-1 inventory
Astrochemical forecasting
Reproducing chemical inventories
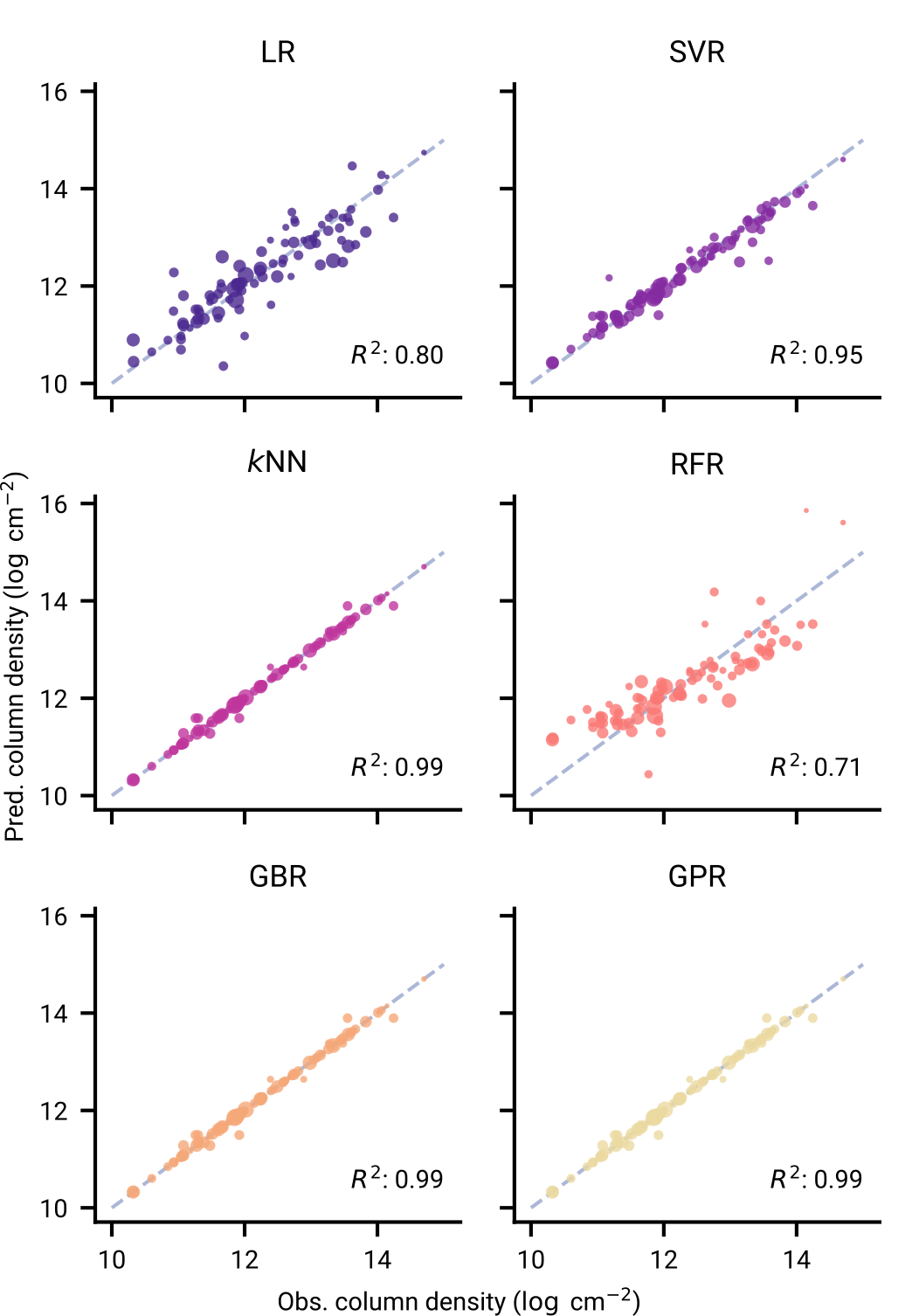
As simple as linear regression to custom kernels in Gaussian Processes
Outperforms conventional chemical models
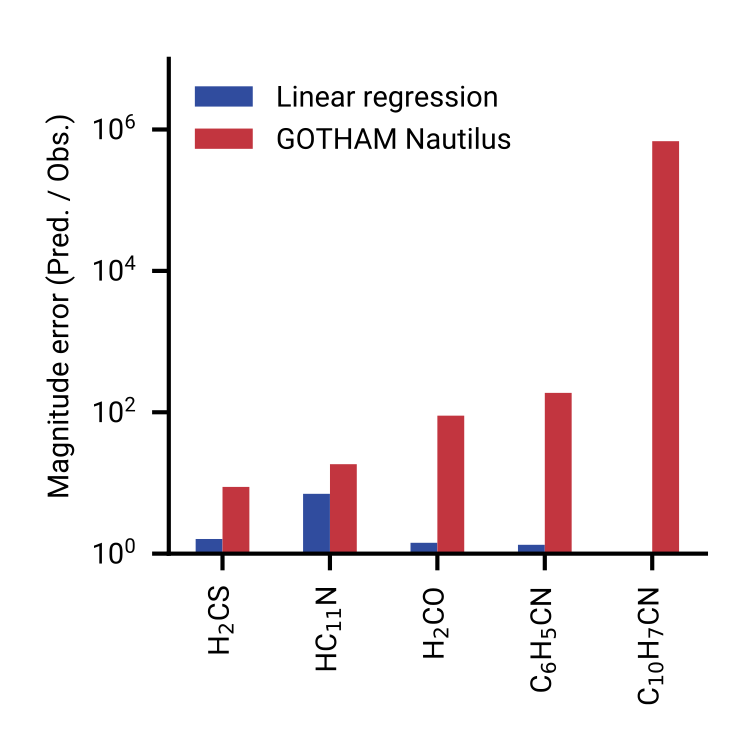
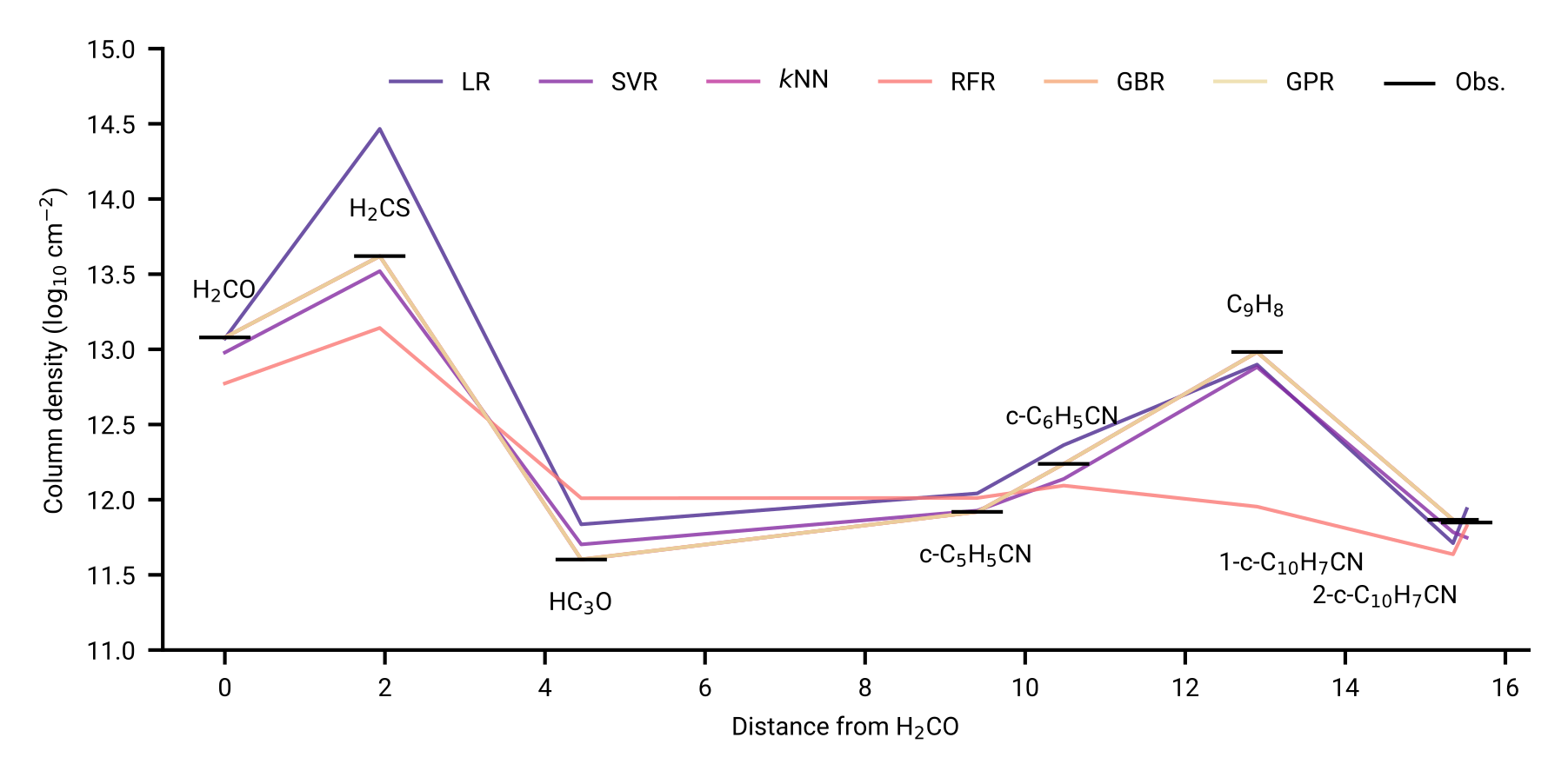
Model performance across chemical complexity
Predicting the abundance of potential molecules
1507 unique molecules identified from 100 nearest neighbors per molecule in TMC-1
17x the number of currently detected species
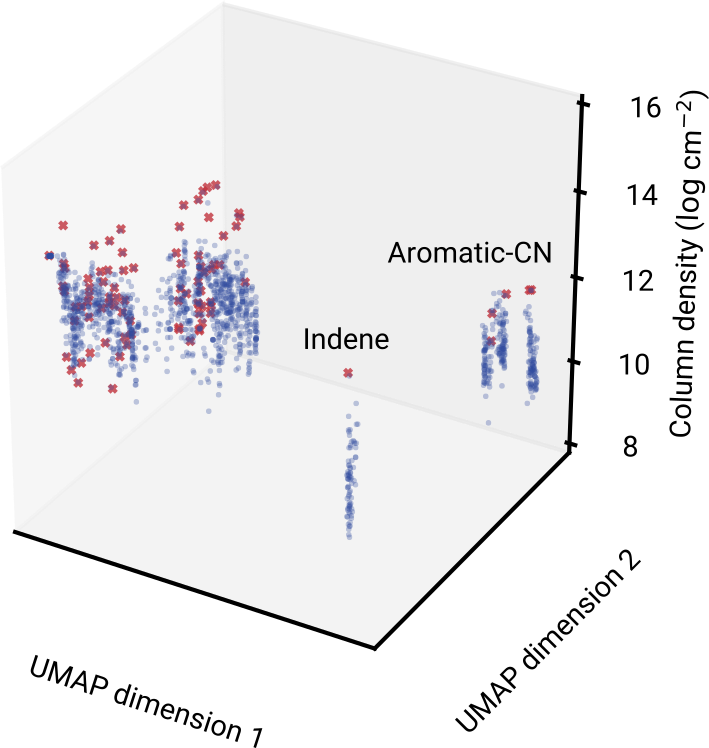
Molecules for laboratory and astronomical study
Out of 1507 molecules...
59% are unsaturated
12% are pure hydrocarbons
43% contain are cyanides
New aromatics up to three rings
Under review—contact me for the full list!
Where to from here?
Apply ML approach to other prototypical sources
Infer abundance of radio-invisible molecules
Guide laboratory/observation/modeling efforts
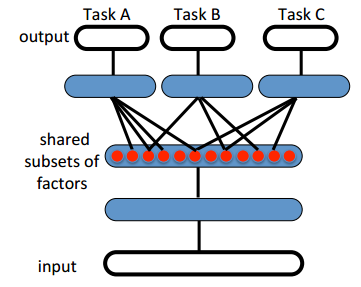
Multi-output models
Acknowledgements


Thank you!
Finding new molecules to study by intuition alone is difficult.
mol2vec brings a new way to study astrochemical inventories.


Use ML to screen molecules
Identify candidates for study and predict their column density
