Unknown Molecule Identification
with Probabilistic Deep Learning and Rotational Spectroscopy
- Kelvin Lee,
- Michael McCarthy
ISMS 2021—Talk FF02
Complex discharge mixtures
Three components of analysis
Line identification
Line assignment
Molecule identification
The most time consuming and ill-defined step!
McCarthy & Lee, JPCA, 2020(124) 3002
Molecule identification
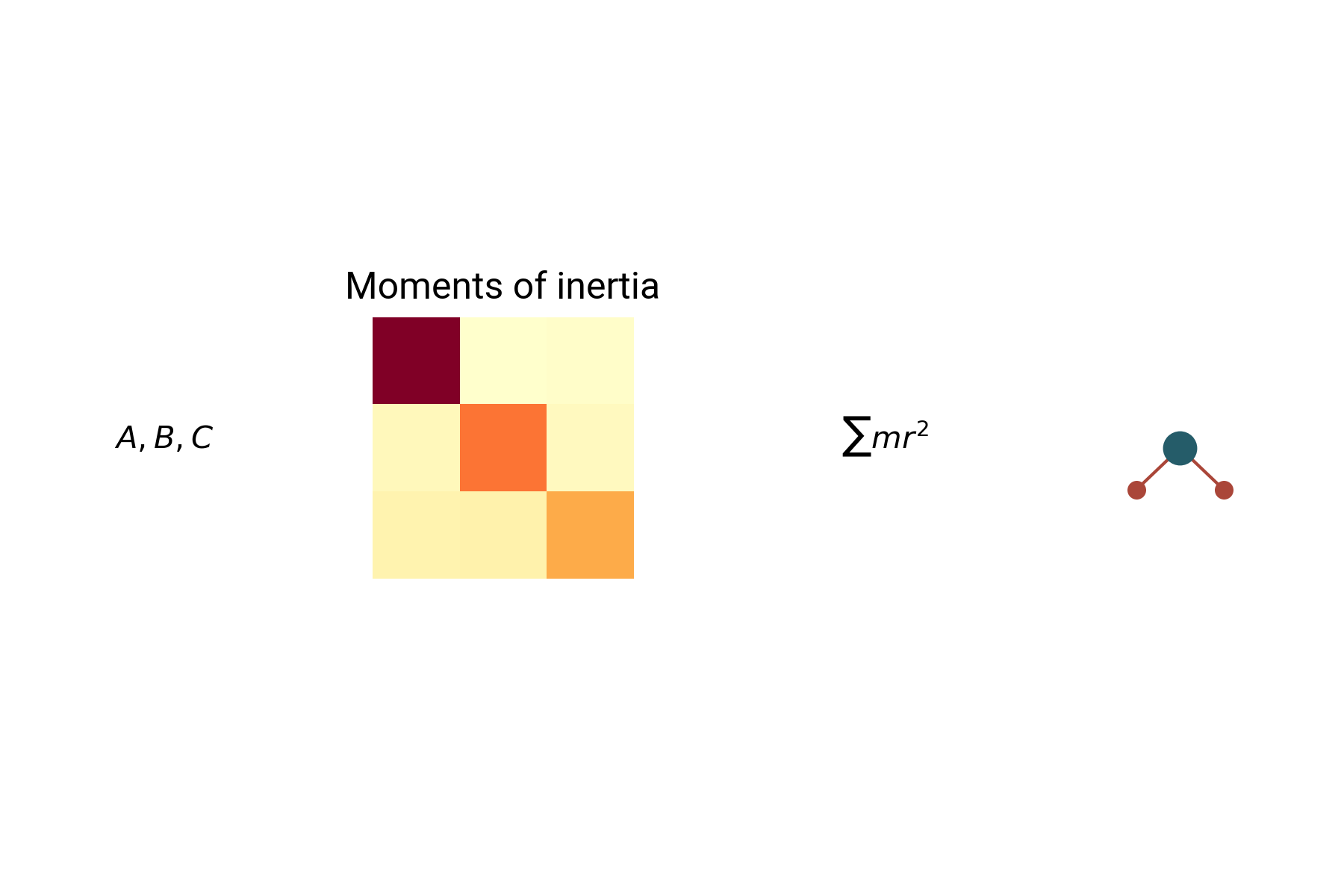
Experimentally determined spectroscopic parameters are uninformative
Mapping constants to structures
Databases
Machine learning
- Mass spectrometry
- Fast—nearest-neighbor lookup
- Uniform accuracy
- Static
- Drug discovery
- Slower, but more modeling flexibility
- Sampling dependent accuracy
- Generative
Deep neural networks
- Universal function approximators
- Parameterize (non)deterministic mapping between constants and features
- Human intuition takes with sufficient information
- Variational inference
Dataset generation
- Molecules systematically generated using Open Molecule Generator
- Up to 2,000 structures per formula, up to $\mathrm{H_{18}C_8O_3N_3}$
- $\omega$B97X-D/6-31G(d) singlet equilibrium structures
- Rotational constants uncertainty used as training augmentation
Structures encoded as Coulomb matrix eigenspectra
$M_{ij} = \begin{cases} 0.5Z^{2.4}_i & \text{for}~i = j\\ \frac{Z_iZ_j}{\vert \mathbf{R}_i - \mathbf{R}_j} & \text{for}~i \neq j\\ \end{cases}$

Ten largest eigenvalues for structurally similar species
No free lunch
Models trained on four subdatasets
Pure hydrocarbons
Oxygen-bearing species
Nitrogen-bearing species
Oxygen/nitrogen-bearing species
Quantitative testing
Use benzene as quantitative testing of model behaviors
Eigenspectrum regression
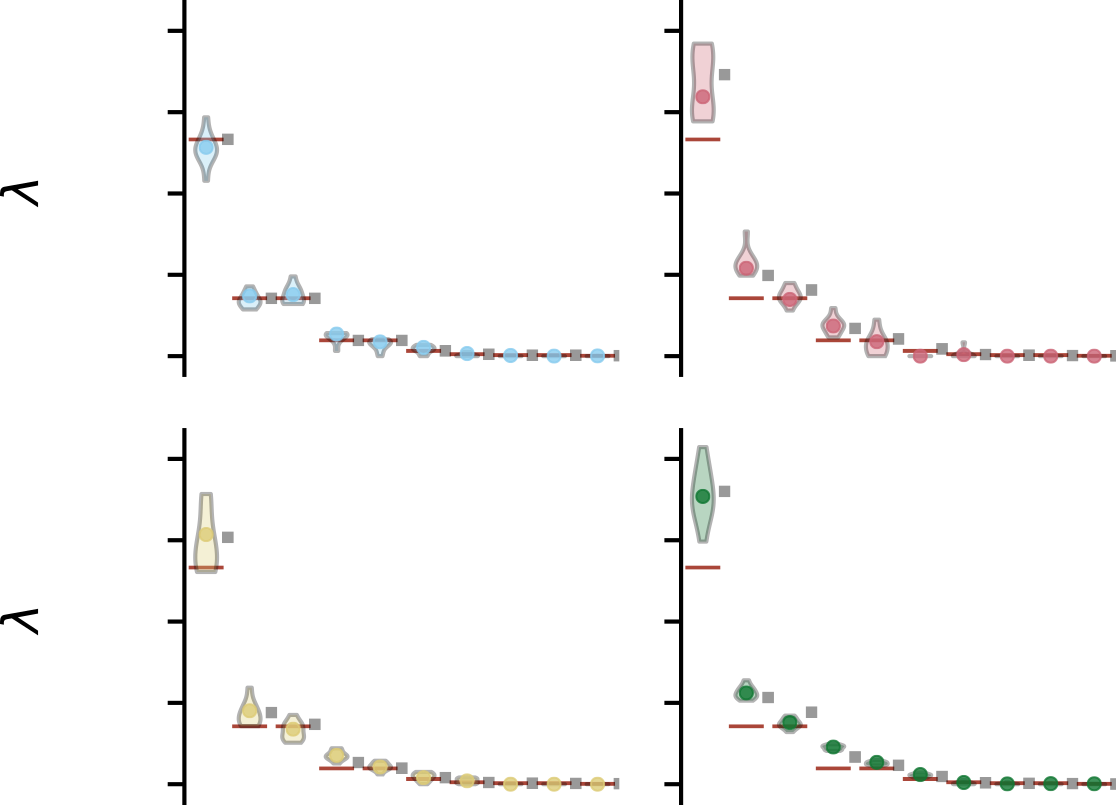
The most critical step—eigenspectra encodes approximate structure and atom composition
What matters most?
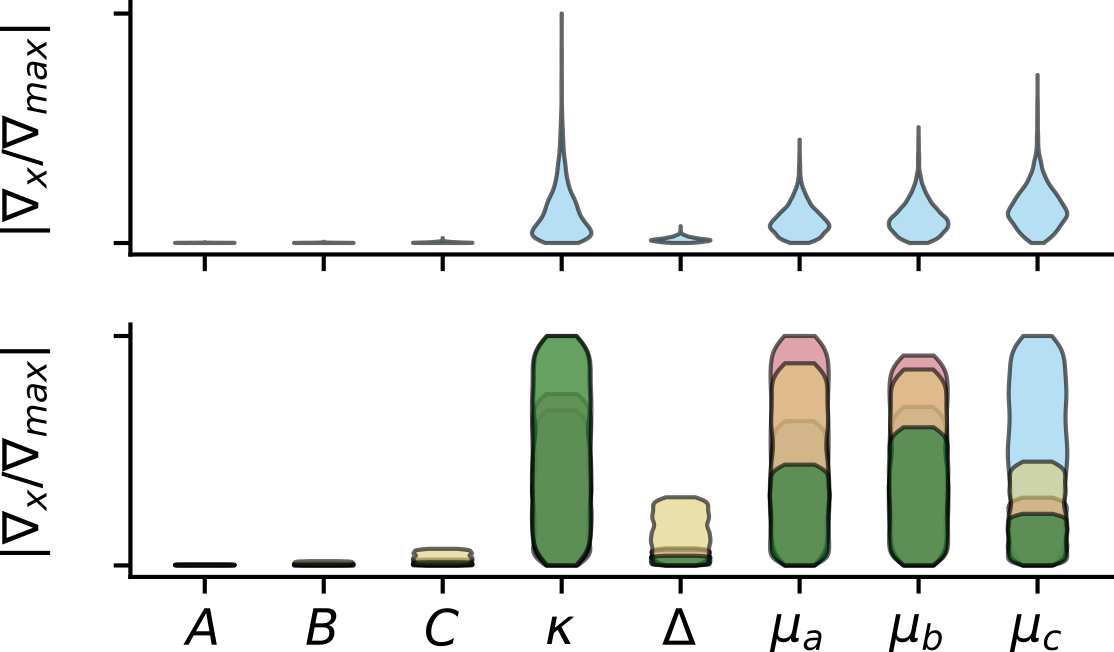
Input gradients indicate $\kappa$ and the dipole moments are most important to the structure
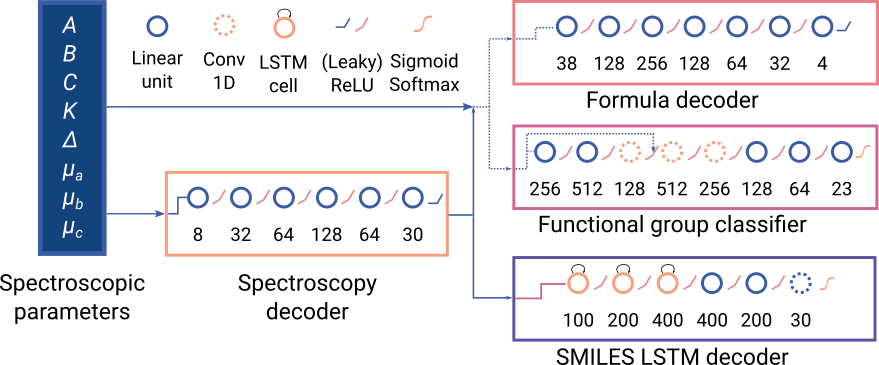
Decoding the eigenspectrum
Extracting identifying information from the Coulomb eigenspectrum encoding
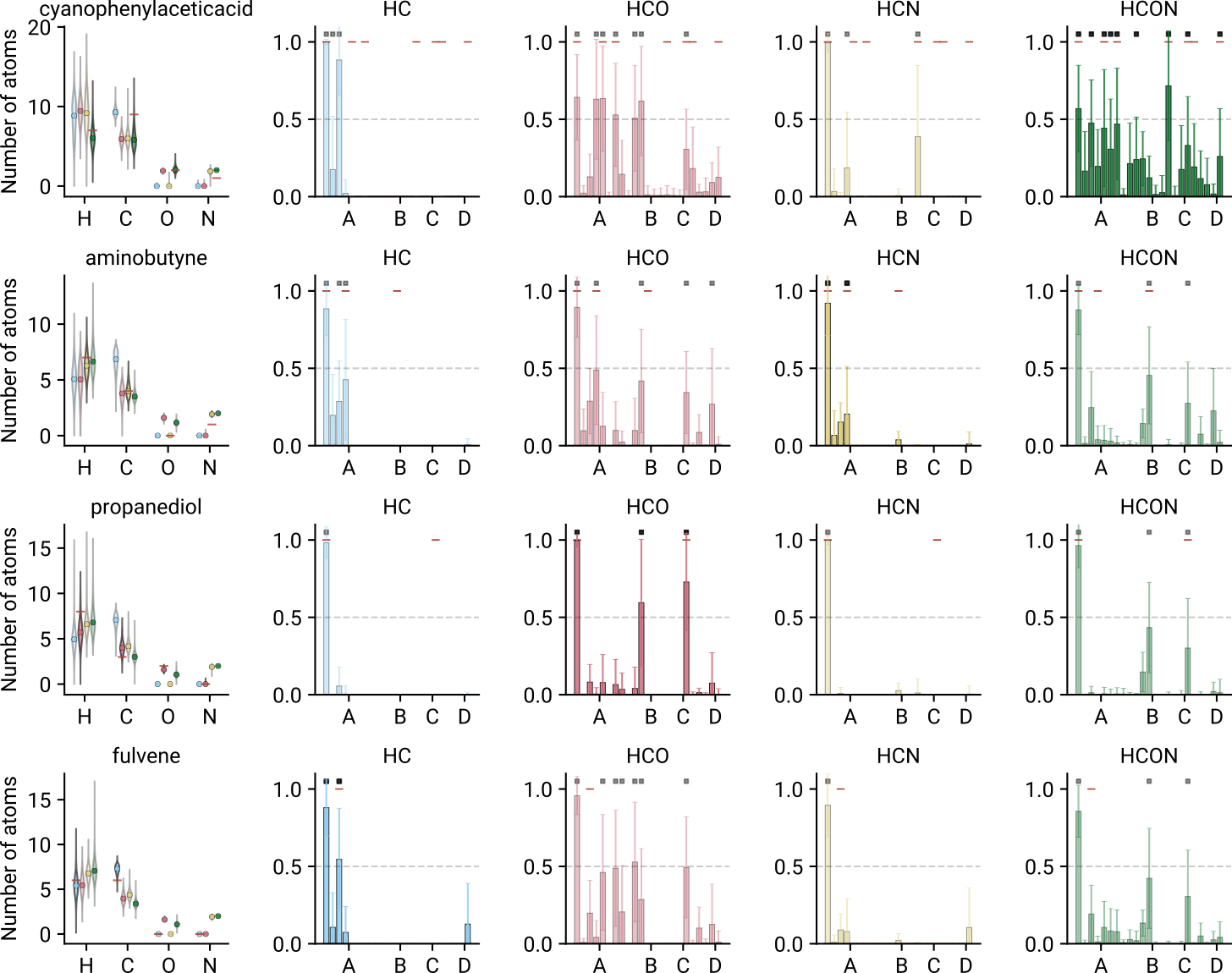
Molecular formula
- Rotational constants Formula
- Hydrcarbon model predicts correct formula within uncertainty
- Remaining models yield roughly the correct mass
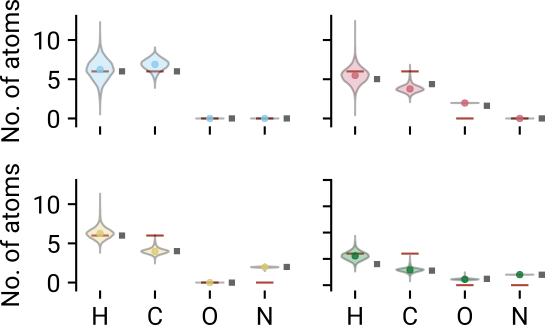
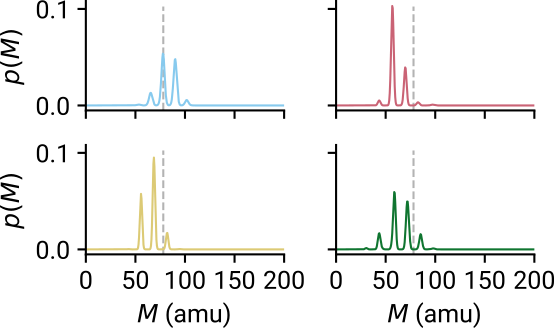
Converted formulae are comparable to mass spectrometry!
Multiclass classification for functional group identification
Intuition from formula + functional group
Areas for improvement
Single model approach
Faster training/inferenceConstants to molecular graph mapping
End-to-end pipeline for molecule identificationAvailable now!
Conclusions
Uncertainty aware deep learning model for molecule identification
Experimentally determinable parameters
Fast, functional interface in PySpecTools
Acknowledgements


Thank you!
Rich complex mixtures provide a wealth of spectroscopic data.

The hard part is identifying completely unknown molecules!

Simple neural networks can identify aspects of the molecule from spectroscopic parameters